“First make it work, then make it faster” — Brian Kerningham
However, if the program takes too long to run, it might not be possible to make it correct. Iterative development breaks down when each iteration takes several hours. Some R code I was working with was supposed to run for a 1000 iterations, but with each iteration taking 75 seconds to run, we couldn’t take the time to let it run to completion. The problem was the continued appending of rows to a data.frame object. Referred to as the Second Circle here. That in turn points to this larger article about problems R programmers find themselves facing. . How does one extricate oneself? Here were my steps.
Table of contents
The Problem
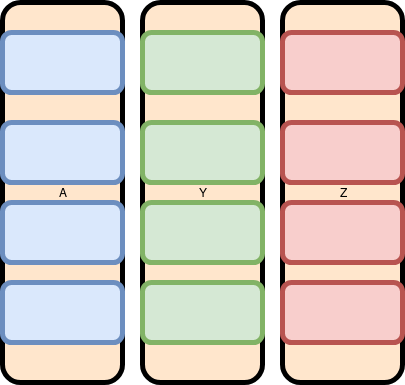
Since Data.frames are organized as a set of parallel vectors, adding a new row requires reallocation and copy. We tend to think of the data like this: Each row contains one entry each of a (A, Y, Z) represented by one of each color Blue, Green, and Red.
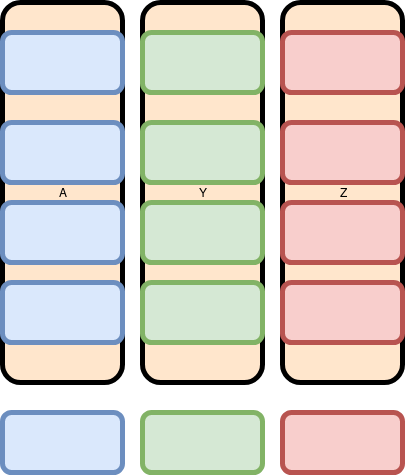
When we want to add a row, we think of it like this:
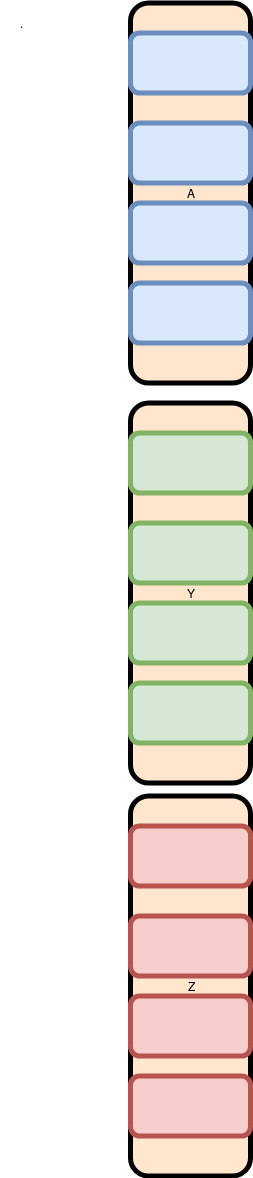
But that is not accurate. Many operations have to be performed on all of the values of a single column. Thus, all the A (Blue) values are kept in contiguous memory. So internally the computer lays them out like this:
And adding a row is more like two inserts and an append:
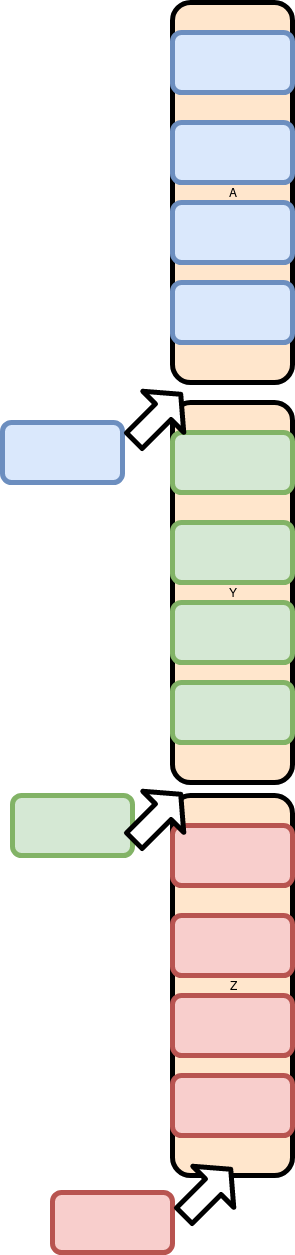
Because all the Data is laid out end to end, the Y and Z columns have to be moved. This is expensive.
The fix is to avoid treating the data as a table until all of the data is generated. When working with individual vectors, we can append values to a single vector with moving the others.
Refactoring The Code
Get it into Git
Even if you are only working with a local repository, Git makes it much easier to revert to a known working version.
create a subdirectory, change into it, and then:
git init git add *.R |
When ever you make a change that you want to keep:
git add *.R git commit -m "Message about the commit." |
Commit early, commit often.
I also put a few entries into .gitignore. Here is what I have:
$ cat .gitignore *~ *Rout .RData .Rhistory rprof.out |
Unit Testing
Setup a unit test or two to confirm the function has not changed. In my case, the code calculates a value called bias_nowt.
[1] “bias_nowt”
[1] 0.2189567
So long as I continue to product this number, my code behaves the same, and I can continue to restructure.
Ideally, I would be checking this value in an automated manner, but this code is not yet in the vicinity of being an R package, and so setting up Unit tests would be more work than called for. It would not take much for me to change my mind on this, though.
Remove commented out code
Make it possible to function inside your code. In the case of the code I am working with, lots of trial and error has lead to much, much commented out code. Removing it reduced the lines of code from 554 to around 300. It makes it easier to not get lost. With old code committed to git, you don’t need to leave it inside the working code.
- Remove commented out Code.
- Run the unit tests.
- Commit to Git.
Profiling
Add a main function
You are going to want to call your code from somewhere else. Take everything that is “top level” and put it into a function.
main(){
#all your top level code
} |
Create a small wrapper that calls the main, like this:
$ cat datagen.R |
Which contains
source("datagen_ipw_instrumented.R")
main() |
and can be run:
R CMD BATCH datagen.R |
Run the unit tests. Commit to Git.
Add a profiling wrapper
$ cat profile-datagen.R |
Which contains
source("datagen_ipw_instrumented.R")
Rprof("rprof.out")
main()
Rprof(NULL)
summaryRprof("rprof.out") |
Use RProf to get actual timing numbers
When I run the code now, I see the times spend in various functions.
At the top of the output, I see:
$by.self
self.time self.pct total.time total.pct
"rbind" 14.64 18.87 40.78 52.57
"[[.data.frame" 4.94 6.37 13.74 17.71 |
And further down
$by.total
total.time total.pct self.time self.pct
"main" 77.58 100.00 0.02 0.03
"FUN" 70.12 90.38 0.28 0.36
"lapply" 70.12 90.38 0.02 0.03
"datagen" 70.04 90.28 1.86 2.40
"rbind" 40.78 52.57 14.64 18.87
"$" 20.06 25.86 3.00 3.87 |
It takes a little bit of analysis of the code to realize that lapply is calling FUN, which is calling datagen, which is calling rbind. The goal is to replace the rbind calls
with something faster.
Start State
Each time the function is called, it initializes some variables, and then uses them to create each of the rows. The initialization code looks like this:
id <- as.numeric(i)
t0 <- 0
U <- runif(1, 0, 1) # Uniformly distributed between 0 and 1
L1 <- rnorm(1, mean=beta1_0+beta1_1*U, sd=sigma)
cumavgL1 <- cumsum(L1)[1]/1 # Calculate cumavg(L1)
L2 <- rbinom(1, 1, plogis(beta2_0+beta2_1*U+beta2_2*L1))
A <- rbinom(1, 1, plogis(alpha0+alpha1*cumavgL1+
alpha2*L2+alpha3*t0))
Y <- rbinom(1, 1, plogis(theta0+theta1*U+theta2*A+theta3*L1+theta4*L2)) |
A few are also passed in as parameters. Later on, it creates a Data.Frame:
temp <- data.frame(Z = Z, id = id, t0 = t0, U = U, L1 = L1, L2 = L2, cavgL1 = cumavgL1,
A = A, Alag1 = Alag1, Y = Y_, wgt_temp = wgt_temp) |
Because these are all scalar values, the data.frame is a single row.
My goal is to replace these with vectors, and call data.frame one with the completed vector.
Introduce Vectors
I added Vectors to the code and insert data into them Parallel to theData.Frame. My naming convention was to create a new variable that matched the original, but with an underbar after it. A Scalar named A would be paired with a Vector named A_.
Code looks like this:
Â
id <- as.numeric(i)
id_ <- c(as.numeric(i))
t0 <- 0
t0_ <- c(0)
U <- runif(1, 0, 1) # Uniformly distributed between 0 and 1
U_ <- c(U)
L1 <- rnorm(1, mean=beta1_0+beta1_1*U, sd=sigma)
L1_ <- c(L1)
cumavgL1 <- cumsum(L1)[1]/1 # Calculate cumavg(L1)
cumavgL1_ <- c(cumavgL1)
L2 <- rbinom(1, 1, plogis(beta2_0+beta2_1*U+beta2_2*L1))
L2_ = c(L2)
A <- rbinom(1, 1, plogis(alpha0+alpha1*cumavgL1+
alpha2*L2+alpha3*t0))
A_ = c(A) |
And so on.
This is a good time to run the unit tests. Nothing should change. Confirm that with the test.
If the run successfully, check the changes in to git. It is an interim change, but one that you might want to roll back to if you mess up the next step.
Add new values to the Vectors
When the loop iterates, it generates a new set of values, and appends to the array. The code looks like this:
# Add data to dataframe
temp <- rbind(temp, c(Z, id, t0, U, L1star, L2star, cumavgL1,
Astar, tail(temp$A, 1), Ystar, NA)) |
I want to make sure all of these values are also captures in the Arrays. I added this code at the base of the loop:
Z_[j] <- Z
id_[j] <- id
t0_[j] <- t0
U_[j] <- U
L1_[j] <- L1star
L2_[j] <- L2star
cumavgL1_[j] <- cumavgL1
A_[j] <- Astar
Alag1_[j] <- tail(A_, 1)
Y_[j] <- Ystar
wgt_temp_[j] <- NA |
Run the unit tests. If the run successfully, check the changes in to git.
Replace data.frame usage
Once all of the Vectors were in place, I started using them in places where the data.frame structure had been used previously. And example of what the code looks like to start:
L1star <- rnorm(1, mean=beta1_0+beta1_1*U+beta1_2*temp$A[j-1]+
beta1_4*cumsum(temp$L1)[j-1]/(j-1)+beta1_5*temp$L2[j-1]+
beta1_7*t0, sd=sigma) |
Note the temp$A[j-1] usage. The variable temp is the data.frame. Since we’ve captured the same data in the Vector, we can replace the data.frame use with the vector A_:
L1star <- rnorm(1, mean=beta1_0+beta1_1*U+beta1_2*A_[j-1]+
beta1_4*cumsum(L1_)[j-1]/(j-1)+beta1_5*L2_[j-1]+
beta1_7*t0, sd=sigma) |
After each of these, rerun the unit tests. You might be tempted to make more changes than this at once and skip running the tests (they take close to 90 seconds at this point) but don’t. You will save more time in the long run triggering your errors right as you make them.
Once all of the temp$ uses have been replaced with Vectors, we can build the return data out of the Vectors instead of using the append. That looks like this:
Replace data.frame return value
temp <- data.frame(Z = Z_, id = id_, t0 = t0_, U = U_, L1 = L1_, L2 = L2_, cavgL1 = cumavgL1_,
A = A_, Alag1 = Alag1_, Y = Y_, wgt_temp = wgt_temp_)
return (temp) |
Run the unit tests. Commit.
You will also want to remove the other call to data.frame and the call to rbind.
Run the unit tests. Commit.
Profile. I get:
$by.total
total.time total.pct self.time self.pct
"main" 28.92 100.00 0.06 0.21
"lapply" 21.42 74.07 0.04 0.14
"FUN" 21.40 74.00 0.04 0.14
"datagen" 21.34 73.79 2.14 7.40
"tail" 8.24 28.49 1.14 3.94 |
and an overall time of
> proc.time() user system elapsed 29.400 0.343 29.734 |
A reduction From 75 seconds to 29.4. Cut the time to execute more than in half. I’m sure we can make additional performance improvements, but we won’t be clipping 35 seconds off the time again.
